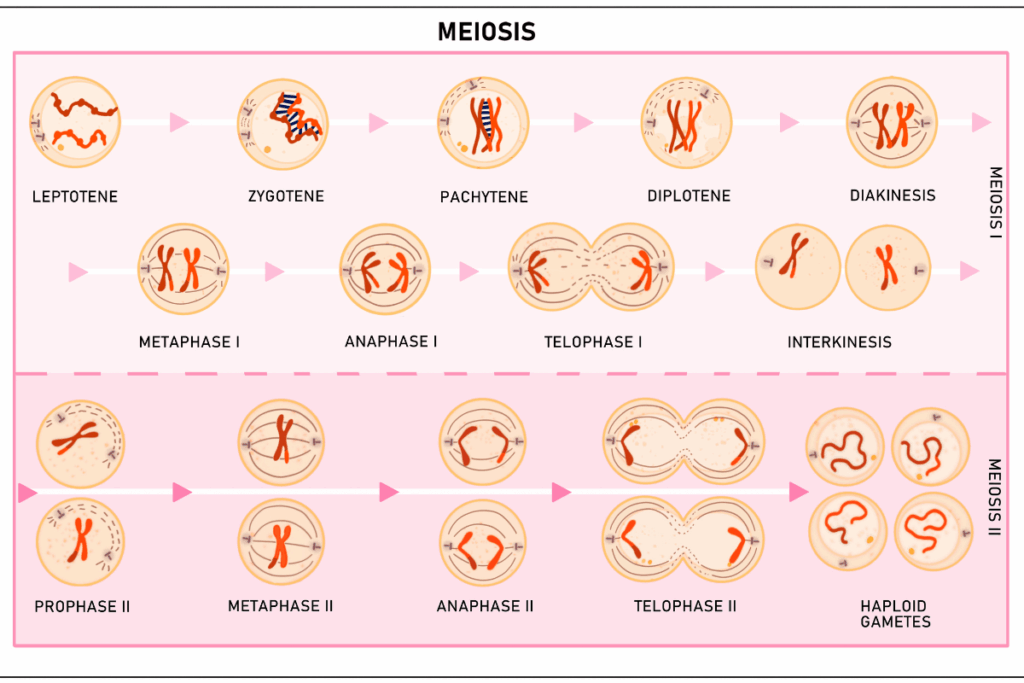
Errors in chromosome segregation during meiosis are a leading cause of early pregnancy loss or first-trimester miscarriage, which in part can be explained by the presence of an abnormal number of chromosomes in embryos, aneuploidy. Although the role of the abnormal crossover recombination in this process has been long established, the shared genetic basis linking recombination patterns to aneuploidy risk was poorly characterized.
A recent large-scale study now provides direct evidence that shared genetic variation in key meiotic genes influences recombination and aneuploidy in humans. This study used pre-implantation genetic testing data of 139,416 in vitro fertilized embryos of 22,850 biological parent pairs, and this was one of the largest datasets to study the meiotic processes taking place in humans. To find out the 3,809,412 crossover events and 92,485 aneuploid chromosomes in the embryos, the researchers employed a new hidden Markov model to trace the transmission of paternal haplotype. Approximately 29.8% of embryos contained at least one aneuploid chromosome, a finding consistent with previous estimates of embryonic loss in early development.
The analysis established that there were fewer crossovers in aneuploid embryos compared to euploid embryos, which supported the argument that crossovers help stabilize chromosome pairing and segregation. Mean maternal and paternal meiosis crossover counts were 49.30 and 31.99, respectively, which are very similar to preceding pedigree-based studies.
Aneuploidies were largely of maternal origin (84,044 maternal and 8,441 paternal), and were highly concentrated on chromosomes 15, 16, 21, and 22. Predictably, the maternal age was strongly correlated with maternal meiotic aneuploidy (0.235, s.e. = 2.19 × 10−3, P < 1 × 10−100), but paternal age was not. It is noteworthy that the authors did not find any correlation between maternal age and maternal crossovers, and therefore, the age-related previously documented recombination changes might be the result of negative selection of aneuploid embryos and not necessarily altered biological rates of crossover.
Genome-wide association analysis studies revealed a strong maternal haplotype in chromosome 22 at the meiosis-specific cohesin gene SMC1B. The most strongly correlated variant, rs6006737, was associated with a higher risk of maternal meiotic aneuploidy (0.066, s.e. = 0.012, P = 2.21 × 10−8) and was also found to be replicated in another test set. In a 40-year-old female, one copy of the risk allele raised the mean risk of aneuploidy by 1.65%, and it was found that the effect was intensified with increased maternal age. Functional studies demonstrated that the risk haplotype is associated with decreased SMC1B expression, in which a non-coding cis-regulatory mechanism, not protein-modifying mutations, is involved. Fine-mapping suggested a variant (rs2272804) at 144 bp upstream of the SMC1B gene transcription start site, which decreases the binding affinity of the transcription factor ATF1 by over threefold, offering a reasonable molecular rationale for the association.
Transcriptome-wide association studies also pointed to C14orf39, which is part of the synaptonemal complex and ubiquitin ligases CCNB1IP1 and RNF212, both of which have been previously identified to regulate meiotic recombination. Some aneuploidy-associated variants were also found to have secondary associations with female age-related phenotypes, such as age at menarche and menopause.
Collectively, these results indicate a common genetic architecture of recombination and aneuploidy, with how common regulatory variation in meiosis-related genes can provide insidious effects on reproductive phenotypes. The study highlights the dual nature of recombination, which enhances genetic diversity and maintains chromosomal stability, providing a new understanding of the biological causes of human fertility and pregnancy loss.
Reference: Carioscia SA, Biddanda A, Starostik MR, et al. Common variation in meiosis genes shapes human recombination and aneuploidy. Nature. 2026. doi:10.1038/s41586-025-09964-2